Single-cell Lung Cancer Atlas
High-resolution single-cell atlas reveals diversity and plasticity of tissue-resident neutrophils in non-small cell lung cancer
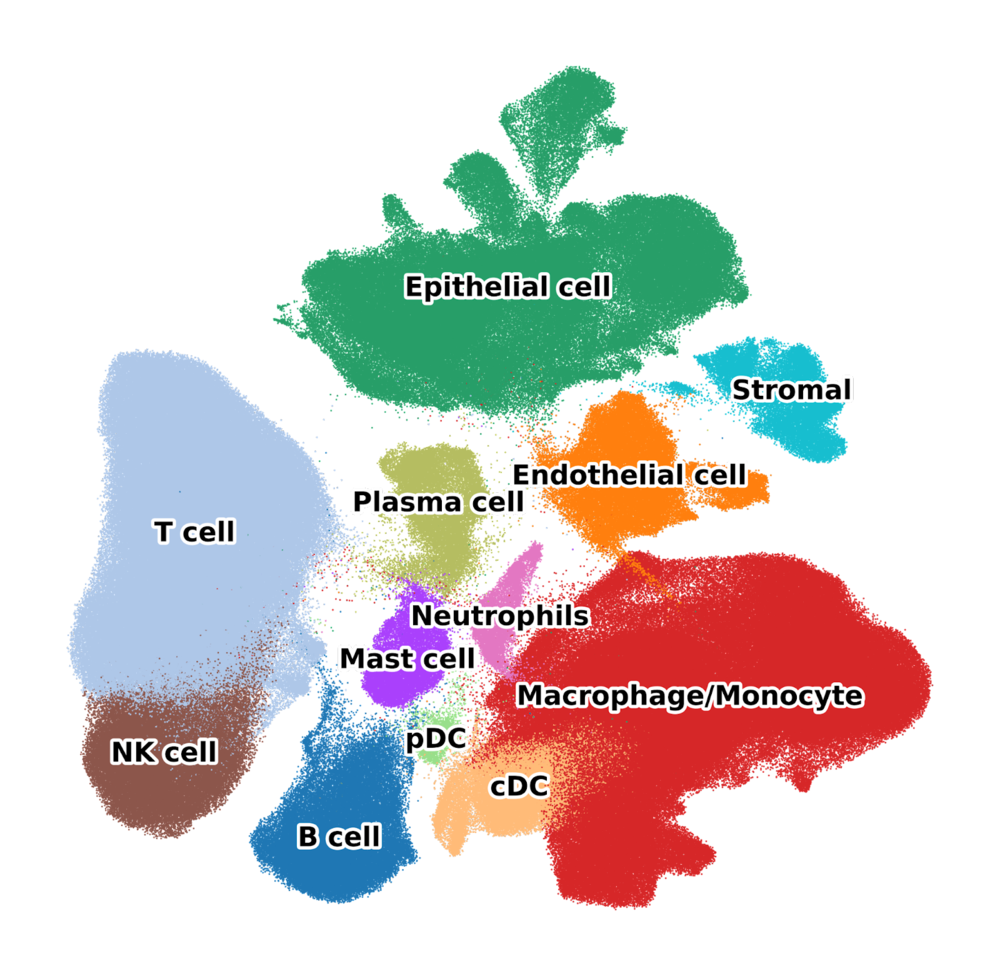
The core atlas was compiled from 21 datasets comprising 505 samples from 298 patients. 38 different cell-types were annotated at different levels of granularity.
By using the transfer learning method scArches another 8 datasets were projected onto the core atlas, resulting in an extended atlas with 1.3 million cells, 556 samples and 318 patients.
CZI cell-x-gene portal
Both core and extended atlas are available on the CZI
cell-x-gene. There, you can
explore the datasets interactively in the browser and
download them as h5ad (scverse) or rds (Seurat) files with standardized
metadata for further analysis.
scArches model
For the core atlas, we also provide a scArches model that allows to project new datasets onto the atlas and automatically annotate cell-types.
Get input data and intermediate results
In addition to the h5ad objects mentioned
above, we provide preprocessed input data, intermediate
results and final results on zenodo. The results were
derived using the source code provided on GitHub and contain
a rendered version of the Jupyter/Rmarkdown notebooks that
were used to generate the figures in the manuscript.
Browse code
All code needed to reproduce the study is wrapped into a nextflow workflow and publicy available from GitHub.
Contact
For reproducibility issues or any other requests regarding single-cell data analysis, please use the issue tracker. For anything else, you can reach out to the corresponding author(s) as indicated in the manuscript.